Let us play a game. Here you have three selections from the recent indexes of three famed journals publishing in the area of Developmental Biology. See if you guess which belongs to which (sure you can put the titles in PubMed and you will find them, but try to do it blindly):
Index 1
1. Large hypomethylated domains serve as strong repressive machinery for key developmental genes in vertebrates
2. Homeotic Function of Drosophila Bithorax-Complex miRNAs Mediates Fertility by Restricting Multiple Hox Genes and TALE Cofactors in the CNS
3. KFoA, a ß-catenin interacting protein linking canonical and non-canonical Wnt signalling during mouse gastrulation and tumorogenesis
4. Actin Cytoskeleton Reorganization by Syk Regulates Fcγ Receptor Responsiveness by Increasing Its Lateral Mobility and Clustering
5. Phosphorylation of CDK2 at threonine 160 regulates meiotic pachytene and diplotene progression in mice
Index 2
1. PIWI homologs mediate Histone H4 mRNA localization to planarian chromatoid bodies
2. Without children is required for Stat-mediated zfh1 transcription and for germline stem cell differentiation
3. Vegfd can compensate for loss of Vegfc in zebrafish facial lymphatic sprouting
4. Gene expression profiling identifies the zinc-finger protein Charlatan as a regulator of intestinal stem cells in Drosophila
5. The DEP domain-containing protein TOE-2 promotes apoptosis in the Q lineage of C. elegans through two distinct mechanisms
Index 3
1. Actin Cytoskeleton Reorganization by Syk Regulates Fcγ Receptor Responsiveness by Increasing Its Lateral Mobility and Clustering
2. RagA, but Not RagB, Is Essential for Embryonic Development and Adult Mice
3. Shh Signaling Protects Atoh1 from Degradation Mediated by the E3 Ubiquitin Ligase Huwe1 in Neural Precursors
4. D6 PROTEIN KINASE Activates Auxin Transport-Dependent Growth and PIN-FORMED Phosphorylation at the Plasma Membrane
5. Developmentally Regulated Elimination of Damaged Nuclei Involves a Chk2-Dependent Mechanism of mRNA Nuclear Retention
It is difficult. It is difficult even to guess which tier (of glamour) do each of them belong to. Rest assured that the abstracts would not help either; they all read the same and yet, when you reveal the brands attached to each of them, turns out that one of them will give more brownie points in job interviews than the other two; and one of them will not give you any points at all -answers later on-. But before getting to the point of this post, apologies to the authors of those papers (if any of them reads this post) for picking them; it is the result of random sampling. They are all good papers and any others would have served equally well. In fact, this is one of the points I want to make: it really doesn’t matter because the intrinsic value of a piece of research does not appear to matter these days.
The first point I want to make concerns the titles, the topics, what is happening to developmental biology. The titles are variations on the theme “gene X/motifM in protein B is required in cell type Y for this/that/ or the otherâ€. Ever since mutant screens took hold of the field, the aim is either to identify new genes and link them to known functions or processes, or to find new functions for known genes (signalling pathways get a prize here). The emerging alternative is those high throughput maps that reflect the toilings of a lot of people, which we are supposed to mine. There is little doubt that this is useful but, is it interesting? Sure, we need to do an inventory of the genes, their products and their modifications and link them to phenotypes but, is there a challenge here? Are there not interesting questions left to answer? I agree that there are technical challenges but, for the most part, these can be dealt with and a product will emerge, a formulaic one. Most papers look like each other, though in the discussion sections there is a bit more latitude in how the results are framed but, for the most part, they are all the same; another piece of a large puzzle, a puzzle whose template we have lost a long time ago. There are problems out there, there are large questions but nobody pushes us to think about them. I have discussed some of these issues before (http://amapress.gen.cam.ac.uk/?p=1094); all I will add here is that slowly, our enquiry is becoming light, unbearably light for those who want to know about and address important issues, or who want to hear those issues. The real questions, that exist, are being diluted by facts.
Thirty years ago we wondered what was the nature of the genes that mediated developmental processes. Many people tackled this question from different perspectives. Some never went anywhere, some did. C elegans and Drosophila brought genes to where chickens and frogs had laid questions and this provided answers to problems. It was (still is, as far as I am concerned) amazing the way it turned out: that the genes we identified in worms and flies had a bearing on vertebrates. But this was, with perspective, satisfying because it meant that there are rules, principles and today, though the indexes of the journals might betray them, we thrive on those principles. With the passing of time, the questions and the answers have become light. Driven by referees, editors, increasing meetings of hype and a general confusion of science with craftsmanship and technology, the lightness of the Science becomes unbearable. An apocryphal comment says that Science might end with us learning more and more about less and less and I am afraid that this is happening in Developmental Biology. If we are not careful, we might end up in the logical conclusion of that path: we shall know all about nothing. What are questions of interest? It will be good to identify them but two of my favourite ones lie in the emerging observation that cells churn out numbers and we do need to understand what those numbers are and where they come from. We need to know how the stochastic molecular world is averaged to the determinism of development and also what is the structure (not the hairballs that people call networks) of the machinery, the molecular devices that mediate the averaging. More on this in the future but here be dragons and perhaps this is why it is easier (but not technically) to carry on with the catalogues.
The reason for all this is that, for the most part the biomedical sciences have become a job which spins out other jobs, particularly in the journal business. I would claim that more than 50% of the publications in the areas of molecular, cell and developmental biology have as their primary aim to serve as career token. Some of them provide information but most of them are just registers of a job done. And here lies the reason why biomedical sciences is becoming unbearably light, because there is less and less hard science in much of what is being produced (read the indexes above). The ‘careerism’ and overcrowding of biomedical sciences is being discussed in several forums. Like many, I hope that we can find ways towards a solution that will restore ‘scientific enquiry’ to the heart of biomedical sciences (at some point I promise to discuss the relationship between the physical and the biomedical sciences which has some interesting lessons for us, biologists). And in this low grade industrial game we are playing, the most important things seems to be not the scientific value of the work but the ranking of the place where the work is published. Yes, I know, this sounds familiar. Time to get to the second point and disclose the journal to which the indexes above belong:
Index number 2 is taken from Development, index number 3 from Developmental Cell and number 1 is a concoction from three journals: Development (title 1), Dev Cell (titles 3, 4) and Developmental Biology (title 5) with a made up paper, did you guess it?, number 2. The point is indeed that they are all the same and that it really doesn’t matter how we distribute them. The titles I like are because they highlight what this journal game is about:
“Vegfd can compensate for loss of Vegfc in zebrafish facial lymphatic sprouting†and “RagA, but Not RagB, Is Essential for Embryonic Development and Adult Miceâ€,
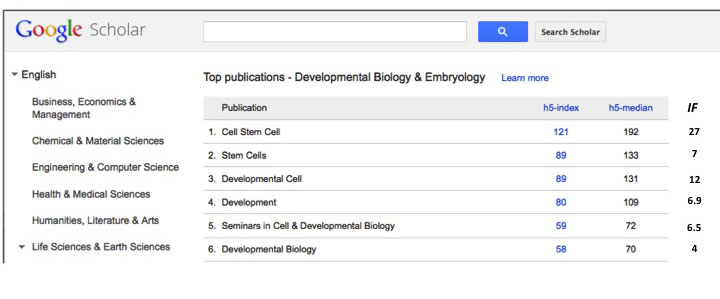
essentially the same issue at stake. Different genes, different organisms, but the same. Almost mirror images of each other and yet, the paper in Dev Cell will give more kudos to the authors. Why? If there was ever a case for SFDORA I think this makes it clearly. It is the same product branded differently but a paper in DevCell will be looked upon as higher quality than on in Development or Dev. Biol. A recent observation in Google Scholar makes the similarity of the two journals more blatant: Dev Cell has an impact factor of 12 (not that great for all their bravado), Development 6.9 and Developmental Biology 4. However, if we looked at the h5-index we get an interesting perspective: Dev Cell’s is 89, Development’s 80 and Dev Biol’s 58. Difference vaporize, particularly between Dev Cell and Development.
One hopes that this will be noted. Not the lack of difference but the inflated nature of some perceptions. In the meantime, a “collective we†still fall for the gimmick of Cell Press (or Nature or Science). Their editors (and publishers), who engineer the game, will tell you that they have the highest standards, particularly in the context here those of Dev Cell. I remember talking to one of the big editors of Cell many years ago, suggesting that I did not like the way they were twisting molecular and developmental biology. This person said to me: ‘we don’t back any horses’. To which I replied that they did not need to, because they bred their own. They have now such a stable that they can organize their own races/conferences around it. It will be claimed that the papers in Dev Cell will have had a more tortuous reviewing process than the ones in Development, which makes them of higher standard, and that some of those in Development or Dev Biol might have been rejected previously from Dev Cell. Some of this is probably true (the rejections, the lengthy pointless review process) but do not be fooled, read the titles, the papers. The quality is the same and the advance that they convey –their scientific content- will also be the same. The difference is the marketing of the product and Cell Press is very good at selling their own. The kudos of a paper in Dev Cell is not based on the rigour, the novelty, the interest or the impact of the paper –as shown above- but by the marketing of the product. And, do not forget that indeed, it is harder to publish there, that the papers in Dev Cell will have more of a spin, that they will have costed more money (in revisions); but nothing else. It is a bubble (remember the tulips in Holland in the XVII century: http://en.wikipedia.org/wiki/Tulip_mania) and we should burst it. Cell Press is general is mostly tulips.
In the end, coming back to the science, none of the journals discussed here address or deal with real problems, with serious questions. This is not for lack of these, it is that the real issues don’t sell (apparently this is what these journals want to do) because they might not have an easy answer, because they cannot solve the problem in a marketable way. What does a journal rather publish: “gene Z controls DNA repair and HXYAc in development and cancer†or “the process of time averaging of gene expression cannot be explained by epigenetics� The good news is that we see less and less screens at the forefront of the journals, the bad news is that this is being substituted by an ever increasing catalogue of more or less interesting but certainly unbearably light assessments of the functions of genes. A lot of work but, for the most part, facts in search of a reason. And most of it driven by journals, with their impact factors relish. Techniques should answer questions not hope for questions.
Solutions? The key lies in the attitude of the current generation of postdocs and graduate students since the future belongs to them. Some things are not going to change but there is something that needs changing: we need to recover the control over what we do, we need to get rid of the tyranny of the journals and the editors. An involuntary collusion of funding bodies and journals has created a very complex atmosphere in the biomedical sciences which has trivialized the science. We need to prise back from them what is ours. SFDORA is one step in that direction and I am constantly surprised how little known it is within the postdoc community. SFDORA is, at the moment the only forum which we can fuel to begin to solve the problem. The other is, of course, a bit more difficult as it relies on thinking what are the real issues in modern developmental biology. But perhaps what I sense as an unbearable lightness in the field is because there is nothing else of interest to be found out…….I don’t think so.

It’s encouraging to hear a senior developmental biologist with these views. I feel there is much more to be learnt about spatiotemporal patterns of development and their regulation beyond what is known. It seems that a lot could be done in putting pieces of what we know already together. There are also numerous opportunities to think outside the square. To hear these viewpoints provides some explanation why there seems to be too few people thinking ahead. I guess the jobs and funding are inside the square, where I’ll undoubtedly have to return to find a job myself. In the meantime, I have a chance to move further up the mountain.
prof premraj pushpakaran writes — 2018 marks the 100th birth year of Edward Butts Lewis!!!