Lists, catalogues and classifications have always been the business of the biological sciences. The nature cabinets of the XVII and XVIII centuries, the collections that occupied much of the XIX century and which fuelled the work of Darwin are good examples of this. Beetles, butterflies, fish, pigeons, plants occupied (and occupy) the time of individuals, often amateurs, interested in Nature. The nature of this enterprise is captured in Umberto Eco’s book “The Infinity of Listsâ€
Lists, catalogues and classifications have always been the business of the biological sciences. The nature cabinets of the XVII and XVIII centuries, the collections that occupied much of the XIX century and which fuelled the work of Darwin are good examples of this. Beetles, butterflies, fish, pigeons, plants occupied (and occupy) the time of individuals, often amateurs, interested in Nature. The nature of this enterprise is captured in Umberto Eco’s book “The Infinity of Listsâ€
When we don’t know the boundaries of what we want to portray, when we don’t know how many things we are talking about (….) when we cannot provide a definition by essence of something and so, to be able to talk about it, to make it comprehensible or in some way perceivable, we list its properties (…………….). We call this representative mode the list, or the catalogue
Indeed: to make something whose limits or meaning we ignore, we make lists, if they are organized according to some criterion (and since Linnaeus but even before, they are) they have the potential to reveal something of the essence of that which is being classified. Physicists and chemists know well how this works: stars, spectra and the elements come to mind. But the level and intricacy of what the biological world offers to the catalogue aficionado is different, probably, boundless. To go back to Eco, it is unclear where the limits of the biological world lie. No wonder E. Rutherford said that one could reduce the sciences to Physics and stamp collecting; he may have had the biological (then natural) sciences in mind and this perhaps is why S. Brenner famously retorted that what Rutherford did not know is that there are some stamps that are worth collecting.
In the History of Science lists have the potential to highlight generalities which allow precise questions to be asked and answered. Physics and Chemistry have been good at reaping the benefits of this activity. In Biology a most famous outcome of this cataloguing is, of course, Darwin’s great work which revealed a principle running through the continuum of transformations that stares from large collections, ordered collections (the word ordered and in what manner the order comes about being important here), of plants and animals. In a different way, the work of Mendel is a culmination of less structured but no less significant collection of lists of the output of many lists; after all, it is seeing patterns in the outcome of crosses of plants that leads to genetics. In all cases the assumption is that if the lists are arranged according to the right criterion, they will reveal an order and, behind that order, some mechanism -in the sense of a causal explanation for a set of observations and not as the usual Figure 7 characteristic of modern biology papers- that will provide an insight into a system. In the end, sometimes, the insight can lead to the manipulation of the system for the benefit of the observer: lists lead to science that leads to engineering which leads to progress.
There is a danger in these lists and it is that they might become an end of themselves. That the scientist becomes a collectionist, forgets Brenner and gives credence to Rutherford. Surely the lists are valuable resources for those that want to ask questions, but the truth is that as we turn into list makers, we can forget that there are questions behind the observations and habit turns us lazy and content in our collecting. Sometimes one feels that this is happening in the biological sciences, that biologists are becoming professional collectionists. There might be a reason for this:  the essence of biological systems is the generation, selection and competitive propagation of novelty and variation. As a result, every species, every genome, every cell in every genome, every organelles in the cell, every protein in the organelle, is subject to this continuous generation of variation, to the exploration of a large space of form and functions. If one assumes that every cell type in an organism is different and that these differences are species specific, one can do a simple calculation: the range of different cell types varies between 3 in a plachozoan to about 1014 in a human and if, as it is currently assumed, there are on the order of 8.5 x 106 organisms on the earth, one could say that there might be on the order of 1020 different cell types to explain (NB this is assuming that all individuals within a species are similar and forfeiting the development of an organism during which large numbers of transient cell types are generated that differ from their final types). This number, 1020 , is already a large number relative to the approximate number of stars, 1012 . It may be small relative to number of atoms, 1080 in the Universe –and one has to remember that atoms need to be proportionately distributed into 117 elements which is where the differences appear i.e all atoms of an element are essentially the same and thus, the 1080 number needs to be tempered by its being bundled up in the abundance of each element. It is here, in this notion of similarity of all the atoms of an element, that the main difference between the biological and physical systems appear. The stars are very similar to each other in composition, and this is why we can study them from a distance by using the spectra. On the other hand, every organism, every cell type in every organism is different, unique. In fact you and I are very similar but our cells in similar places in similar organs are likely to be different. Enter DNA, which is the way to explain uniqueness in Biology: if we accept, as we must, that every cell type responds to a ‘transcriptional code’ of sorts, and we focus just in humans with our approximate 20,000 genes (I am not interested in philosophical discussions of what is a gene and hope that you and I will agree that this is a lower bound), simple calculations allow for 220000 combinations, to account for those 1014 different cells (and don’t forget those developmental intermediates). If you throw this number into your calculator, it will be confused as it will approach infinity. Of course, the toilings of Natural Selection ensure that only part of that repertoire is used but still, the number is large and dwarfs anything the inorganic world can produce. Surely we are stardust, like the moons of Jupiter, but DNA and RNA have found a way to turn that dust into a creative material device.
Where am I going with this? Over the last few years technical developments have allowed us to peer into single cells at the level of their transcriptional complement and, with increasing accuracy, at the level of their genomes. The observation is that even within what histologically is a (one) cell type, there is a great deal of heterogeneity. It is difficult to silence the genome, and we are learning that cells –particularly in development- are exploring their transcriptional space in a dynamic manner. The result is that within an organism much of that space of 220000 combinations is likely to be explored and much of it represented. The technical developments are allowing increasing volume and accuracy in the observation of this process (gene expression at the level of single cells) and of the delivery of these results. In consequence this creates interesting challenges for classifying, for making lists, which are taken on by groups of computational biologists whose interests lie in dealing with complexity rather than in understanding its meaning. Meetings are held on the subject of gene expression at the level of single cells and while at the moment the possibilities lie in honing our ability to describe the expression patterns of single cells and of characterizing the genomes of cells in tumours, the holy grail on the horizon is the analysis of epigenetic marks at the level of single cells and the ambition of getting the genome, epigenome, transcriptome and proteome in single cells. Our infatuation with these techniques, what it reveals and the possibilities associated with it are powerful and thus reviewers and editors lurk in the background to ask you for a single cell analysis of your favourite system, if everything else has failed to hamper the publication of your work. But, at the moment, it is also expensive and begs the question of where does it lead to? What is the meaning of this work? Are we paying lip service to Rutherford?
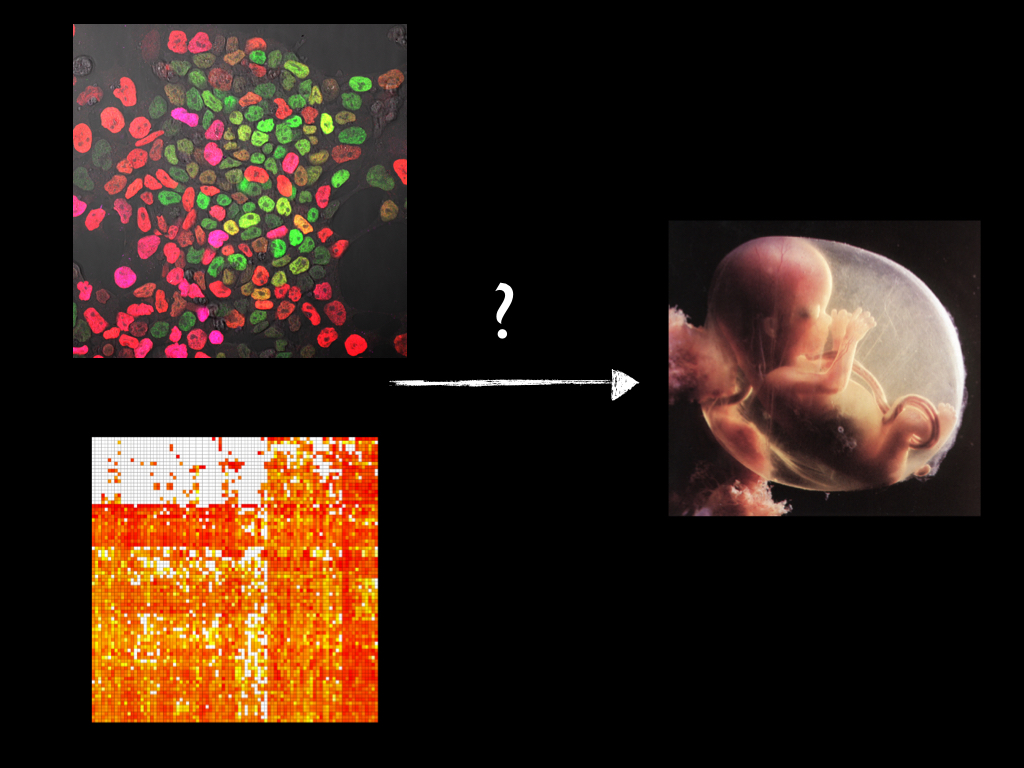 The analysis of single cell gene expression can have -and sometimes had- an impact in three areas of Biological research: Cancer Biology, Immunology and my area of interest, Developmental Biology, which aims to understand how an organism builds itself. In all cases, single cell analysis allows the identification of ‘rare cells’ which sometimes have a function and sometimes, they don’t. The issue is that more often than not and in the best tradition of Biology, these studies reveal the temptation of collecting data under the banner of its ‘importance’ without realizing that we have fallen to a fad, that cataloguing has taken precedence over understanding. The description of a biological process demands a link between a cellular and a genetic description of the process and there is little doubt that the arrival of single cell transcriptomics and associated techniques, particularly single cell lineage tracing, has revolutionized the field. However we should be careful not to be swayed by the collectionist syndrome and remember that behind the data there are questions and that if we cannot see them, we should acknowledge that. We should not confuse cataloguing and collecting with Science. In some ways there is no great difference between beetles and genes, and we might be developing a XXI cabinet of genes and cells. It might require more challenging techniques than those collections of the past but there is no difference between collecting one or the other. Already papers in journals tend to be divided into two: either analysis of gene X in tissue Y in organism Z, or increasingly, single cell analysis of process W in organism Z. And in the best tradition of classical Systems Biology, one hopes that in the analysis of the data, the question and the answer will emerge at the same time as one stares in hope at the data.
The analysis of single cell gene expression can have -and sometimes had- an impact in three areas of Biological research: Cancer Biology, Immunology and my area of interest, Developmental Biology, which aims to understand how an organism builds itself. In all cases, single cell analysis allows the identification of ‘rare cells’ which sometimes have a function and sometimes, they don’t. The issue is that more often than not and in the best tradition of Biology, these studies reveal the temptation of collecting data under the banner of its ‘importance’ without realizing that we have fallen to a fad, that cataloguing has taken precedence over understanding. The description of a biological process demands a link between a cellular and a genetic description of the process and there is little doubt that the arrival of single cell transcriptomics and associated techniques, particularly single cell lineage tracing, has revolutionized the field. However we should be careful not to be swayed by the collectionist syndrome and remember that behind the data there are questions and that if we cannot see them, we should acknowledge that. We should not confuse cataloguing and collecting with Science. In some ways there is no great difference between beetles and genes, and we might be developing a XXI cabinet of genes and cells. It might require more challenging techniques than those collections of the past but there is no difference between collecting one or the other. Already papers in journals tend to be divided into two: either analysis of gene X in tissue Y in organism Z, or increasingly, single cell analysis of process W in organism Z. And in the best tradition of classical Systems Biology, one hopes that in the analysis of the data, the question and the answer will emerge at the same time as one stares in hope at the data.
Single cell analysis of expression is the epitome of this strange hypothesis-free science that is often hailed in reviews and social media. We are in the midst of it. Slowly we fool ourselves that large data and cataloguing will lead us to the essence of a process, that it will allow us to talk about something that we cannot define. And while it is true that Biology has a habit of revealing principles from lists I cannot help but thinking that with this trend of hoarding data, we are losing perspective of the processes that still need addressing. It would be good if, as R. Feynman said, we don’t confuse naming something with understanding something. Developmental Biology in particular, is losing itself in this naming game and single cell analysis will –unless checked- provide the ultimate distraction from questions that are there but we are too…..may I say ‘lazy’? to ask. We should not forget that there are things to explain, that cataloguing is a way to answer, and sometimes to unlock, those ‘things’ but also that we need to make an effort to search for them.
The allure of the information that can be gathered in one of those experiments is enormous but one needs to remember that in addition to being expensive and data rich, it needs to be insightful. The difference between a collectionist and a scientist should lie not in the ability to make observations but in the ability of the second to use the observations to answer specific questions about Nature. Biological systems have a boundless ability to generate (constrained) variability and it seems to me that the challenge is to understand the nature of the machine –for it is a machine- that generates, processes and uses that variability, written in that tape that is the DNA, interpreted by the transcriptional machinery and supervised by Natural Selection. It is the process, not its output, that needs to be explained. Questions are cheaper than data gathering but good questions are hard to come by.
Epilogue
One of the most disturbing aspects of the current trend in the single cell field is the lack of cross reference or discussion of the data. Often the same system is surveyed in more than one paper without any reference to the other, related, pieces of work or even, on occasion, to the general problem. While this is in keeping with the current trend in the biological sciences in which the publication rather than the finding is what matters, it is no less disturbing. If we do not get hold of the boundless nature of that data by using questions to clean it up and thus reveal what is good and bad data, we shall do a disservice to the system that puts up the money for that research and, more importantly, to Science itself. Surely, there is meta-analysis, Darwin’s great work can be construed as a meta-analysis- but nowadays, often this is done not so much with a question in mind but with the idea of multiplying the data-analytical power. The boast tends to be not in what has been learnt but in how large the data set is. And in the end, the danger is that, increasingly, what we do is expensive collecting; XXI century cabinets of genomic data, without a good reason, without a good question –which exists. We seem to have relinquished our ability to interpret what we observe and lost our interest in asking questions because, I agree, it is easier to order and catalogue this diversity that we call Biology. Still, the issue buzzes in the back of my mind: there are questions, important questions, to be asked and….all that data!