Outline of a proposal for the inner structure of the cell: an interactive project.
Here I would like to put forward a seed for a framework to think about the ‘inner structure of the cell†which should help framing the outcome of genetic screens. S. Brenner has talked about the inner representation of an organism when referring to the manner in which its different parts, its development are encoded in the genome. I would argue that in order for this notion to be useful and to be useful in the use of Genetics to unravel it (see the companion piece ), we need to tackle the functional organization of the cell; its physiology. I want to stress that this is a seed that it is hoped to grow through interactions and comments from all of you. If you have any and want to participate with information and ideas, write to ama11@hermes.cam.ac.uk. Maybe we can work together in an interactive project.
Instead of a multiply connected interactome view, it is suggested that the cell is organized hierarchically with four levels that, at a higher level, integrate development with the biology of the cell (Figs 1-7).
Figure 1. At the core of the system there is a relatively small number of Gene Regulatory Networks (GRNs) organized into network motifs, configuring a collection of Information Processing Units (IPUs) that interact with each other through some of their component elements in a cell type specific manner. The networks will be cell type or developmental stage specific and many will share some of the elements. These GRNs are of the kind that have been discussed by U. Alon, M. Elowitz and colleagues; they have the intrinsic property of generating temporal patterns of activity and, through interconnections and the feedback of downstream elements, to generate programmes of gene expression. In some cases the networks are cell type specific (e.g mesoderm, endoderm, neural…….) in others the same elements are connected in different manners in different cell types. Thus, cell types, high in a developmental hierarchy, are specified by specific networks linked through particular connectivities of existing networks. This structure allows for these IPUs to rewire themselves within and between each other. In all cases the connections have variable strengths, are dynamic, and are subject to biochemical fluctuations (see Trott et al. 2012 for an example of these notions at work).
Figure 2. The core of IPUs is encapsulated in a signalling environment made up of Signal Transduction Networks (STNs) which has a more loose structure but which nevertheless is constrained (in terms of interactions). The activity of this STN layer looks at the collection of IUPs from which it also receives inputs to form a Core Information Processing Center (CIPC), which is likely to be cell type specific. With cell type not meaning ‘terminally differentiated†cell type. The interaction between STNs and IPUs endows individual cells with specific phenotypes; the fluctuations derived from the chemical nature of the connectivity generates expression heterogeneities, which have been described and which become very important in transitions between cell states or cell fates. Signalling contains two kinds of functional units: some that control the connectivity between and within the individual GRNs, and those that regulate the strength of the interactions between the nodes (in the past we have suggested that Wnt and Notch signalling are involved in this activity see Martinez Arias and Hayward, 2006). The interactions between these two levels (IUPs and STNs) are, like those between the elements of the IUPs, plastic and cell type specific but probably affect ALL cell types i.e. the CIPC is a dynamic structure.
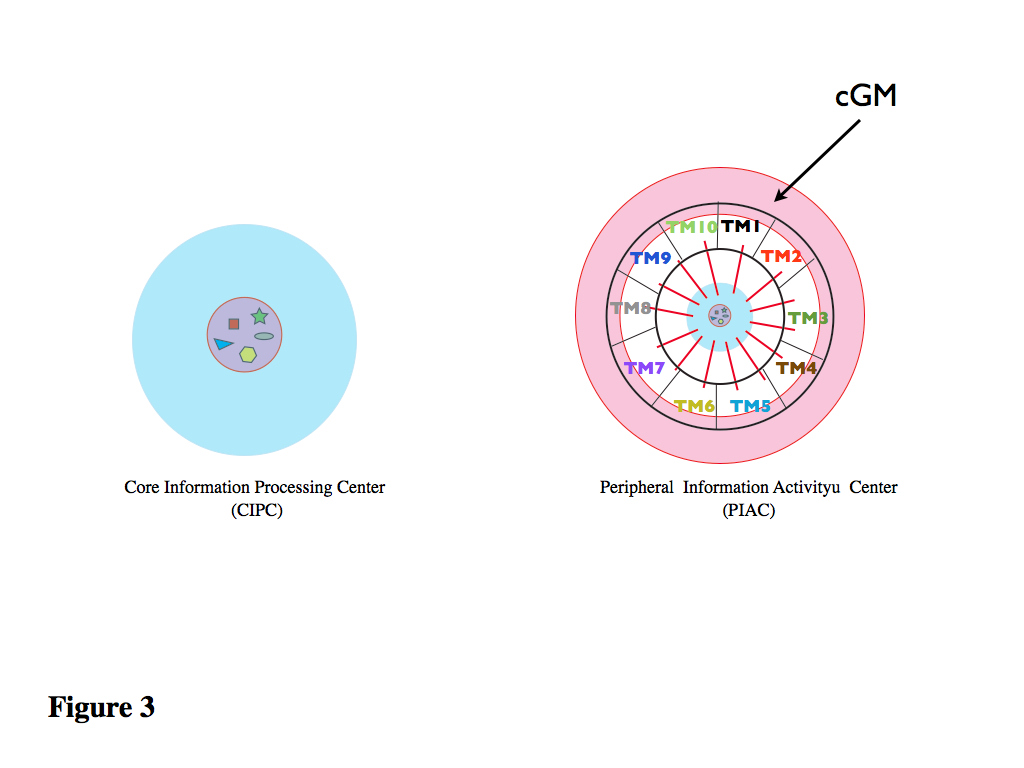
Figure 3. There is now a discontinuity between this Central Core (CIPC) and the next level (Peripheral Information Activity Center) which is made up of two components: one not cell specific composed of the basic cellular General Machinery (cGM) and some circuits (TM for Transducer Modules, protein circuits in this case) which act as interphases between the environment, the CIPC and the PIAC.
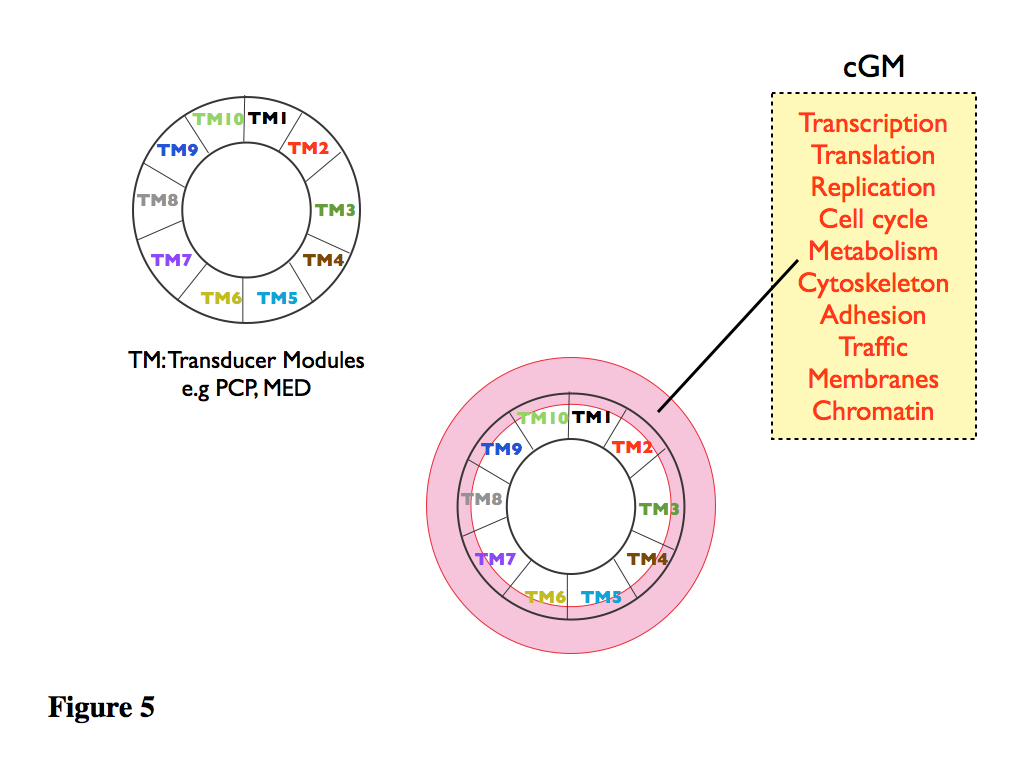
Figure 4, 5. At the heart of the outer layer, there is a collection of modules (TM) whose function is to take information from the outside (or the CIPC inside) and link it to module specific cellular General Machinery (cell cycle, cytoskeleton, traffic…..see Fig.4 for a list). Thus, the TMs lead to a cell type integration of the cGM into cell type specific activities. An example what is meant by this is provided by the module known as Planar Cell Polarity (PCP) pathway. In this view, PCP is a molecular device used to translate extracellular cues (chemical and mechanical) into the cytoskeleton, and is cell type specific i.e its components are shuffled and organized in a cell type specific manner. Equivalent modules can be found for transcription (in particular certain chromatin remodellers and the Mediator complex are likely to mediate this function), traffic, adhesion, cell cycle…the staples of the cells. The prediction from this speculation is that there will be other modules in addition to PCP performing equivalent functions. Furthermore, it is likely (and there is already evidence for this) that some of the elements of the PCP module will bridge with adhesion and traffic. Each TM being associated with a function. A second important function of these TM modules is to channel information between cGM and the central CIPC core and this is achieved through its links, direct or indirect, with the signalling space (Fig. 5). These outer layers (PIAC) have a high degree of redundancy and plasticity derived from extensive element sharing.
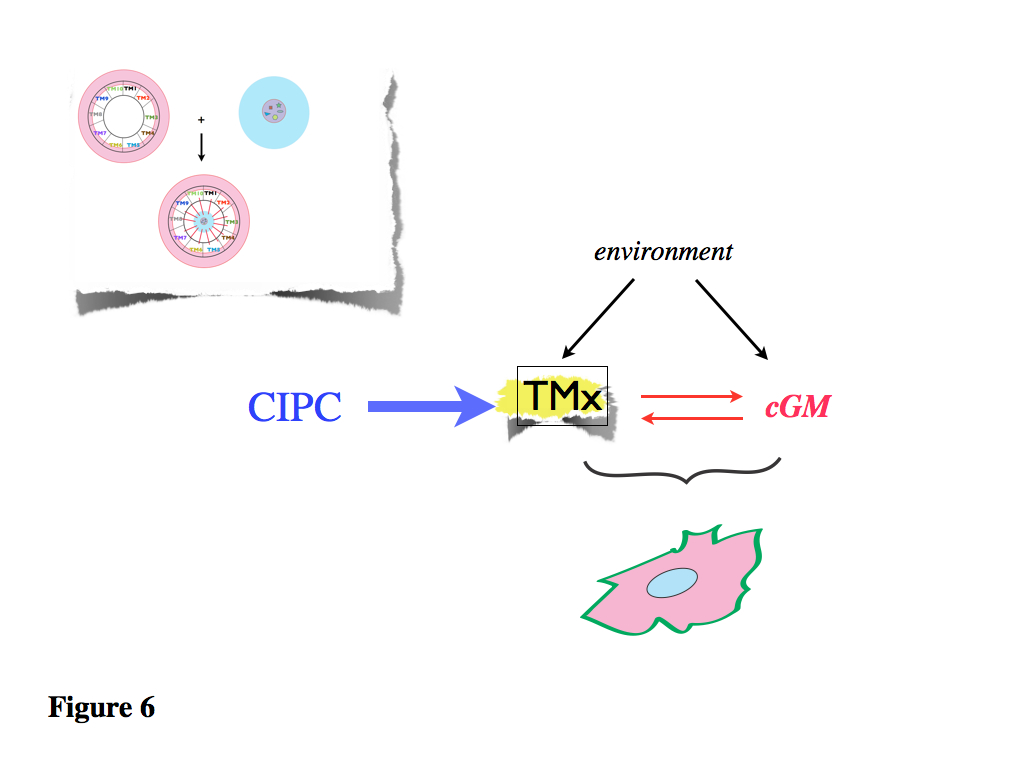
Figure 6. Thus, The TMs are at the heart of the functional definition of a cell type or state, as indicated in this diagram.
Figure 7. This highly speculative organization of the inner structure of the cell could account for the outcome of genetic screens. Thus, one would say that the early genetic screens in the 80s and 90s identified many of the components of CIPC, because they were looking at global compounded phenotypes (body plan, presence or absence of organs or structures, loss of cell identities early in development which would have knock on effects on the general structure of the organism e.g mutations in major signalling pathways or common elements of GRNs). The screens did not target connectivity or structure thus we really do not know much about this. We know something about cell type composition i.e. what elements correspond to which lineage or which cell, but not structure. As the screens get refined they target elements of the higher levels, the TMxs or the results of losing elements in T and GM. There is a very high degree of genetic redundancy in both levels, associated with the plasticity of the system and therefore, the phenotypes can be more subtle.
Problems and issues for discussion: This is very general but it might be useful as a start. Perhaps it could be a good idea to apply it to a specific problem, cell type or developmental system. But its value is that it should be helpful in explaining the outcome of genetic experiments in a general manner.
References
Martinez Arias, A. and Hayward, P. (2006) Filtering transcriptional noise during development: concepts and mechanisms. Nat. Rev. Genet. 7(1):34-44.
Trott, J., Hayashi, S., Surani, A., Babu, M. and Martinez Arias, A. (2012) Dissecting ensemble networks in ES cell populations reveals microheterogeneity underlying pluripotency. Mol Biosyst. 8(3):744-752.





I would talk of “organisation of” or better “information flow in” cells rather than “inner structure”, since structure and physiology seem not to be well represented in the models so far.
This is interesting and simplifies thinking of incoming signals. Now, could you think of similar schemes to explain cellular responses to signalling? My impression is that we know much less about the logic of these. We tend to represent the response of the cell to signalling as a simple arrow (activation of X gene) or a multitude of parallel arrows (as in figure 4 above). These different responses must also be organised as interacting, perhaps hierarchical, logical modules. To me, the lack of such a model for cellular responses is what is stopping us from understanding any complex developmental process at the cellular level. It appears like a mess once you hit ‘cytoskeleton’ for example.