Once upon time….somebody said that Genetics is Ariadne’s thread of Biology, the only way to guide us out of the labyrinth that is a biological problem. Nowhere has this been more true than in the analysis of development, the processes underlying the emergence of an organism. In this spirit, it is the systematic application of genetic analysis to Drosophila melanogaster and Caenorhabditis elegans spearheaded a deluge of information that started modern ‘Developmental Biology’ (the study of the dynamics of pattern and form in embryos, as opposed to ‘Embryology’, the detailed description of the different stages associated with embryos). The success of these two invertebrates took its time to permeate the mammalian embryo. There are good reasons for this. Two possibly important ones come to mind: the challenge that is to study an embryo that develops inside the mother and, not unrelated, the difficulty of applying Genetics to such an object. In retrospect we can see that most of the mutations that have given us insights into developmental events are embryonic lethal and it has been the pathological, or if you don’t want to sound so dramatic say phenotypic, analysis of those mutations that has yielded a view of the process. Notwithstanding husbandry and breeding details, the mammalian embryo, small and hidden in the confines of the uterus, is not an ideal target of systematic screens (the mammalian genetics papers) but with patience and focus, Genetics was applied and the usual combination of serendipity and method have yielded their fruits.
It could be said that there are many kinds of Ariadne’s threads. For example, mammals have an interesting aspect of their make up that is not a bad one into the labyrinth and that, in my view, has not been exploited as much as it might: haploinsufficiencies. Dominant phenotypes caused by the loss of one allele that are related to the function of a gene and it is with a haploinsufficiency that mammalian Developmental Biology started with the discovery of the mutation that lends its name to the gene Brachyury. In some ways the story (or the path if you want to stick to the mythology) of Brachyury is a story which highlights the highs and the lows of the connection between Genetics and Development, that reveals how easy it is to be distracted by the underlying complexity of biological processes, how molecular biology can simplify things but also how we should not be complacent and forget that genes are not causes but simply tools and elements to understand a process. It is also an example, as it unfolded, of the power of the blend of Genetics and Embryology that forms the core of XX century Developmental Biology. Also, perhaps surprisingly to some, Brachyury paved the way for the genetic analysis of development as it was the first mutation (genes are related to mutations but are not the same) associated with a developmental defect, many years before Poulson’s work with Drosophila Notch (1).
 We do know today that Brachyury, also known as T or T/Bra, is a gene that encodes a transcription factor of a family called the T-family because of their structural relatedness, that play crucial roles in early development (2). We also know that it has a central role in the processes of gastrulation and axial extension in chordates and that its existence extends to invertebrates where it has a variety of roles in early patterning of embryos (3).
We do know today that Brachyury, also known as T or T/Bra, is a gene that encodes a transcription factor of a family called the T-family because of their structural relatedness, that play crucial roles in early development (2). We also know that it has a central role in the processes of gastrulation and axial extension in chordates and that its existence extends to invertebrates where it has a variety of roles in early patterning of embryos (3).
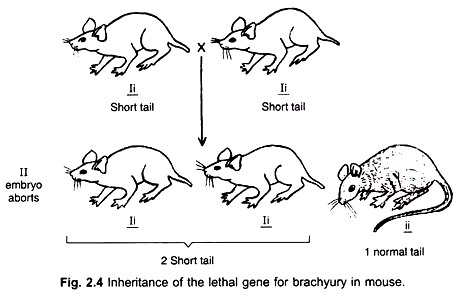
The origin of Brachyury lies in the 1920s Paris where Nadine Dobrovolskaia-Zavadskaia, a Russian emigrée from the revolution, was working at the Institute for Radium (an interaction between the Curie Laboratory and the Institut Pasteur) looking for the ability of X-rays to induce mutations in mammals (4). In the course of a screen involving 3000 crosses of mice across three or four generations she found two mutations that bred true, one of them she called T. The mutation had a dominant phenotype reflected in the length of the tail, hence the name T for taillessness (the capital for the dominant phenotype) but was, also, embryonic lethal (5).
An obsession with T starts and, in parallel with the discovery of recessive alleles, t, and interacting mutations (in laboratory and wild mice), descriptions of the phenotypes of different genetic combinations suggest an involvement of T, whatever it was, with the development of spinal cord and of the vertebral column (6-8 and then, if you want to follow on, read the more general accounts in refs 9 and 10). One thing is to describe a mutant phenotype –a forensic exercise- and a different one to wonder whether it tells something general about the connection between genes and developmental processes; I am, not sure that we have yet resolved this problem. It was, probably, Salome Gluecksohn-Schoenheimer who began to do the second through her analysis of T and t mutants (8, 10). There had been descriptions of the defects of the mutants (6-8) but she went further. Having worked in Spemann’s laboratory (11) the defects in the development of the notochord, neural tube and somites associated with T mutants, made her think of the effects of the organizer and of a possible relationship between T and its activity. These thoughts were no doubt encouraged by her discussions with, particularly C. Waddington, and the parallels she drew with mutations affecting the patterning of the Drosophila wing, suggested that T was saying something of how genes related to developmental processes (10, 11). But these were early days to see, let alone study, this relationship. Ariadne’s thread was in a badly tangled ball and the genetics of T did not help. Mutants with T like phenotype, some allelic, some not but which acted as modifiers were isolated in wild and laboratory strains, and their relationship to the original T mutant proved genetically complex (see brief discussion of these matters in refs 9, 10 and, more extensively, 12). The notion that these genetic interactions were linked to the function of T, led to a labyrinthine situation.
The problem was, and still is, that the genetic analysis of a biological process has its limitations; it is a bit like palm reading. It requires interpretation and, in the case of complex processes –like Development- it is not the way to unravel Mechanism. To quote a very prescient thought from Gluecksohn-Schoenheimer  “A mutation that causes a certain malformation as the result of a developmental disturbance carries out an “experiment†in the embryo by interfering with the normal development at a certain point. By studying the details of the disturbed development it may be possible to learn something about the results of the “experiment†carried out by the gene. However to discover anything about the nature of the action of the gene is a much harder task. It is necessary for this purpose to be able to trace back all the results of the action to certain original causes†(8). Furthermore, Genetics for its own sake (the analysis of the complexity of interactions, alleles, pseudoalleles, complex complementation associated with the breeding of a trait) can intervene, capture our imagination and lead us astray. A bit like maths without Physics; elegant and fun but lacking bite. However, Genetics was what there was and the genetics of T proved a bit of a tangle (12), particularly when it came to the question of the link of the mutants (and at the bottom the wild type gene) to the generation of the embryo.
The question of what was the relationship between genes and development was an important issue in the 60s and the 70s. It would emerge from the molecular and cellular analysis of T, and in parallel of the bithorax complex (BX-C) in Drosophila by Ed Lewis. How genes controlled development did not look a simple affair at the time. Being interested in the topic as a graduate student in the late 70s and the early 80s I remember reading the seminal –but somewhat arcane- 1978 paper by Lewis on the BX-C (13) and a profoundly puzzling one on T (14). I must admit at having been simultaneously fascinated and befuddled by both but particularly by the mouse one. The complexity and the challenge to unravel them was part of the excitement to solve the puzzle. The Drosophila case seemed easier to unpack, perhaps; and it was. The reason probably lies in the rather Cartesian organization of the embryo –which was rather well laid out at the time- and the linear manner in which events unfold that allowed a rather rapid connection between genes and specific developmental events. It has interested me recently, how much hard work was going on to outline the battleground to apply Genetics to mammalian embryos when the buzz was about the genes of Drosophila. But T was not forgotten as it held a key to how mammalian embryos were built and therefore needed to be addressed.
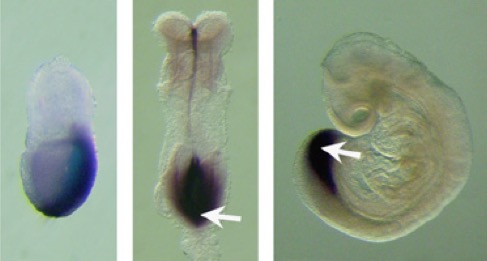 From here, the story gathers pace and the riddles find their solutions. And so it was that the era of molecular cloning clarified matters. In the 80s the systematic application of DNA cloning and analysis to the harvest of mutants screens from Drosophila and C. elegans, paved the way for similar work in other organisms, particularly mammals, and started to lift the fog that Genetics has laid on T. The BX-C turned out to encode three transcription factors and a complex regulatory region (15,16), while T/Bra encoded one transcription factor (17,18). The nature of the devilish genetics of T/t still lingered in the background but the brutal simplicity of molecular analysis delivered its verdict: T was, IS, a transcription factor expressed in the notochord as well as the progenitors of the spinal cord and the mesoderm (see Figure). The question then was, not how this related to the complex genetic analysis revealed by the multiple alleles and crosses –this is, apparently, still work in progress- but how this related to the activity of the protein, encoded by the gene and revealed by its loss of function. And this was just the beginning (in developmental biology, knowing what kind of protein is encoded by a gene –that we have come to know from a mutant phenotype- often opens more questions than it answers). The question was now the original one and again Gluecksohn-Schoenheimer, presciently posed it;  in 1938 thinking about how Spemann’s group approached the problems associated with embryonic development she mused about how in an ideal world to approach mammalian development: ‘The events that take place in the development of the mammalian embryo have not been subjected to an extensive causal analysis so far. The reasons for this are to be found mainly in the lack of suitable methods. It is not possible yet to use transplantation, isolation or vital staining techniques on mammalian embryos as they have been used on amphibian embryos. In the course of time it probably will be possible to analyze the mammalian embryo by transplantation and isolation just as thoroughly as has been done with the amphibian. For the present, however, the experimenter is not able “to take an active part in the course of events that take place during the embryogeny of the mammalian embryo,†nor “to alter the course of events at a chosen point in a chosen manner and draw conclusions on their relations from the resulting changes.†(Spemann 1936.)’ (8). An amazing paragraph for the time. She was more right than perhaps she thought and the molecular biology of T was going to pay high dividends that would make her musings true. The 80s gold rush of gene cloning revealed that T/Bra was conserved across phyla and its discovery in Xenopus led to a fruitful link with mesoderm induction that Jim Smith and his colleagues pursued in an enlightening manner for many years, establishing how T/Bra worked at the molecular level and established a connection between T/Bra and gastrulation (see e.g 19, 20). In parallel, Rosa Beddington began to apply the techniques that were being developed to study mouse development to the molecular insights and reagents thus unravelling the connections between T/Bra and mammalian development that lurked behind the genetics for so long (21-23). This work was soon picked up by one of her collaborators, Valerie Wilson, who has been pursuing the intricacies of the relationship between T/Bra and the mammalian body plan for the last many years answering many of the questions that had been posed by the early Genetics of the mutant. There is still much to do because, thought we have learnt much, the question of the relationship between gene and effect, mutant and phenotype, remains. But now we have the tools and the framework to try to answer it. We just need not be distracted by the ease to gather facts and remember the questions.
From here, the story gathers pace and the riddles find their solutions. And so it was that the era of molecular cloning clarified matters. In the 80s the systematic application of DNA cloning and analysis to the harvest of mutants screens from Drosophila and C. elegans, paved the way for similar work in other organisms, particularly mammals, and started to lift the fog that Genetics has laid on T. The BX-C turned out to encode three transcription factors and a complex regulatory region (15,16), while T/Bra encoded one transcription factor (17,18). The nature of the devilish genetics of T/t still lingered in the background but the brutal simplicity of molecular analysis delivered its verdict: T was, IS, a transcription factor expressed in the notochord as well as the progenitors of the spinal cord and the mesoderm (see Figure). The question then was, not how this related to the complex genetic analysis revealed by the multiple alleles and crosses –this is, apparently, still work in progress- but how this related to the activity of the protein, encoded by the gene and revealed by its loss of function. And this was just the beginning (in developmental biology, knowing what kind of protein is encoded by a gene –that we have come to know from a mutant phenotype- often opens more questions than it answers). The question was now the original one and again Gluecksohn-Schoenheimer, presciently posed it;  in 1938 thinking about how Spemann’s group approached the problems associated with embryonic development she mused about how in an ideal world to approach mammalian development: ‘The events that take place in the development of the mammalian embryo have not been subjected to an extensive causal analysis so far. The reasons for this are to be found mainly in the lack of suitable methods. It is not possible yet to use transplantation, isolation or vital staining techniques on mammalian embryos as they have been used on amphibian embryos. In the course of time it probably will be possible to analyze the mammalian embryo by transplantation and isolation just as thoroughly as has been done with the amphibian. For the present, however, the experimenter is not able “to take an active part in the course of events that take place during the embryogeny of the mammalian embryo,†nor “to alter the course of events at a chosen point in a chosen manner and draw conclusions on their relations from the resulting changes.†(Spemann 1936.)’ (8). An amazing paragraph for the time. She was more right than perhaps she thought and the molecular biology of T was going to pay high dividends that would make her musings true. The 80s gold rush of gene cloning revealed that T/Bra was conserved across phyla and its discovery in Xenopus led to a fruitful link with mesoderm induction that Jim Smith and his colleagues pursued in an enlightening manner for many years, establishing how T/Bra worked at the molecular level and established a connection between T/Bra and gastrulation (see e.g 19, 20). In parallel, Rosa Beddington began to apply the techniques that were being developed to study mouse development to the molecular insights and reagents thus unravelling the connections between T/Bra and mammalian development that lurked behind the genetics for so long (21-23). This work was soon picked up by one of her collaborators, Valerie Wilson, who has been pursuing the intricacies of the relationship between T/Bra and the mammalian body plan for the last many years answering many of the questions that had been posed by the early Genetics of the mutant. There is still much to do because, thought we have learnt much, the question of the relationship between gene and effect, mutant and phenotype, remains. But now we have the tools and the framework to try to answer it. We just need not be distracted by the ease to gather facts and remember the questions.
The story of T/Bra is a good example of the way in which the blending of Genetics. Embryology and Molecular Biology have enlightened the relationship between genes and development. History can be anecdotal but it is also informative. The recent history of developmental biology is very focused on Drosophila and C. elegans and it may come as a surprise to many that T/Bra, as a question and as a reality, was there before Bicoid and Wnt and Notch, highlighting the questions that needed an answer. History, in this case, also highlights the perils of the purely Genetic analysis of a biological process and the need to remember that Genetics is a language, a formal language, to ask questions. In the analysis of Development, it leads us to the elements of the system but might not be the element of choice to see how they come together to make an organism. The position of Genetics to Biology is a bit like Mathematics to Physics: a language that allows one to formulate a question in formal terms which then provides a machinery to work towards the answer but the output of this operation needs to be interpreted.
The history of Brachyury also has an interesting element in that it highlights the important contribution of women to the field; most of the important breakthroughs and insights in the story come from women: Nadine Dobrovolskaia-Zavadskaia, Salome Gluecksohn-Schoenheimer, Virginia Papaioannou, Rosa Beddington, Val Wilson. An influence worth emphasizing as we celebrate Women in Science week.
In the end, the tale continues. Rather than the English and “they lived happily ever after’, we could quote the French “ils vécurent heureux et eurent beaucoup d’enfants” (they lived ever happily after and had a lot of kids), the kids being the myriad of questions that have been raised by the great discoveries about T over the last twenty years. Discoveries which open the door to answer to the questions that T/Bra leads us into: about Development, Genetics, Evolution, about Stem Cell biology and, in the near future, of the engineering of living systems.
NB One appreciates that this piece just skims through the surface of the story and its implications. Still, one hopes that this will inform at some level and, also, encourage thinking about the connections between Genetics and Developmental processes. I am grateful to Peter Baillie-Johnson for the suggestion of the title. The image on the genetics of T is from: http://www.biologydiscussion.com/gene/genes-types/genes-types-top-6-types-of-genes-genetics/67413
References
1. Poulson, D. 1940. The effects of certain X-chromosome deficiencies on the embryonic development of Drosophila melanogaster. J Exp Zool 83: 271–325.
2. Papaioannou VE. (2014) The T-box family: emerging roles in development, stem cells and cancer. Development 141, 3819-3833.
3. Technau, U. Brachyury, the blastopore and the evolution of the mesoderm. BioEssays 23, 788-794
4. Korzh, V. and Grunwald, D. (2001) Nadine Dobrovolskia-Zavadskaia and the dawn of developmental genetics. Â Bioessays 23, 365-371.
5. Dobrovolskia-Zavadskaia, N., 1927 Sur la mortification spontane´e de la queue che la souris nouveau-ne´e et sur l’existence d’un caracte`re (facteur) he´re´ditaire “non viable.†C. R. Seances Soc. Biol. Fil. 97: 114–116.
6. Dobrovolskia-Zavadskaia, N, Kobozieff N, and Veretennikoff S. Etude morphologique et genetique de la brachyourie chez les descendants de souris a testicules irradies. Arch de Zool Exp 1934;76:249±358.
7. Chesley P. (1935) Development of the short-tailed mutant in the house mouse. J Exp Zool 1935;70:429±459.
8. Gluecksohn-Schoenheimer, S. (1938) The development of two tailless mutants in the house mouse. Genetics 23: 573–584.
9. Pappaioannou, V. (1999)The ascendency of developmental genetics, or how the T complex educated a generation of developmental biologists. Genetics.151. 421-425.
10. Gluecksohn-Schoenheimer S. (1989) In praise of complexity Genetics 122, 721-725.
11. Gluecksohn-Schoenheimer S. (1992) The causal analysis of development in the past half century: a personal. Development 1992 Supplement
12. Silver L.M. (1985) Mouse t haplotypes. Annu. Rev. Genet. 19, 179-208.
13. Lewis, EB (1978) A gene complex controlling segmentation in Drosophila. Nature 276, 565-570.
14. Artzt, K., McCormick, P. and Bennett, D. (1982) Gene mapping within the T/t complex of the mouse. I. t-lethal genes are nonallelic. Cell 28: 463–470.
15. Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. (1983) Science 221, 23-29.
16. Gehring, WJ (1992) The homeobox in perspective Trends in Biochem. Sci. 17, 277-280.
17. Hermann, B. G., S. Labiet, A. Poustka, T. King and H. Lehrach, 1990 Cloning of the T gene required in mesoderm formation in the mouse. Nature 343: 617–622.
18. Kispert A, Koschorz B, Herrmann BG. (1995) The T- protein encoded by Brachyury is a tissue specific transcription factor. EMBO J. 14, 4763-4772.
19. Smith JC, Price BM, Green JB, Weigel D, Herrmann BG. (1991) Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 67, 79-87.
20. Saka Y, Tada M, Smith JC. (2000) A screen for targets of the Xenopus T-box gene X-bra. Mech Dev. 93, 27-39.
21. Beddington RS, Rashbass P, Wilson V. (1992) Brachyury, a gene affecting mouse gastrulation and early organogenesis. Dev Suppl. 1992:157-65.
22. Wilson V, Rashbass P, Beddington RS. (1993) Chimeric analysis of T (Brachyury) gene function. Development. 1993 Apr;117(4):1321-31.
23. Rashbass P, Cooke LA, Herrmann BG, Beddington RS. (1991) A cell autonomous function of Brachyury in T/T embryonic stem cell chimeras. Nature 353, 348-351.

Lovely writeup!