 A meeting was held recently at the Pasteur Institute on the topic “Engineering the embryo: beyond Systems Biologyâ€. The event brought to my mind a question I pose to the final year undergraduate class: how should we approach a biological problem? like physicists or like engineers?
A meeting was held recently at the Pasteur Institute on the topic “Engineering the embryo: beyond Systems Biologyâ€. The event brought to my mind a question I pose to the final year undergraduate class: how should we approach a biological problem? like physicists or like engineers?
The relationship between Physics and Biology has a long and very distinguished history, strewn with technical contributions that have often changed the direction and pace of biological research. Microscopy and X-ray crystallography would not have happened without the intervention of the physicists. To see this you don’t need to go further than the 2014 Nobel prize to E. Betzig, S. Hell and W. Moerner for the deep developments in superresolution techniques that are having a most dramatic impact in cell and developmental biology. However, there are also profound and long lasting conceptual contributions. Neurobiology has benefitted enormously from the input of physicists and molecular biology was shaped by Schrodinger’s “What is life?†and Max Delbruck’s Phage School. More recently, over the last twenty years, a cadre of young physicists have led a renaissance of the relationship between Physics and Biology. Single molecule and single cell techniques have opened up our eyes to the statistical processes underlying molecular and cellular Biology and taught us the beginning of how to deal with them. For those of us who have been lucky to be part of this feast, it has been both fun and insightful. If you have missed it, I suggest you catch up.
The fact that a biological system integrates multiple variables was never in doubt but it has been our ability to access those variables that has changed the game. Faced with a deluge of information of specific processes, we have learnt to measure and to use models to understand those measurements and, in turn, perform precise experiments. In many ways we are at the beginning of this game but the input of the physical sciences forged over the last ten years has already left an important imprint permeating much of forefront biological research. There is much to do and we now know how. Why then mulling over engineering?
The word ‘engineering’ has many meanings but, in general, evokes images of machines and blueprints. If you belong to the last century, like some of us do, you might think of steam engines, bridges, chemical plants, of belts and braces. If you belong to this century you might think of jets, computer chips, speed trains and clean technology, of electronics. Engineering is, in many ways, applied Physics. It is about toiling with materials to do something useful in a reproducible manner. Where a physicist uses an equation to understand/explain a process, an engineer uses it to explore a practical solution to a problem. Where a physicist uses a phase space to explore the behaviour of a system, an engineer will look at it seeking the domain that might work for a specific process in the real world. A phase space may often be the end game of the physicists work, for the engineer it is the starting point. While much of engineering deals with the physical world, there have been many inroads into Biology and, as in the physical world, there are many kinds of engineering in Biology: fermenters, biochemical reactors, bacterial circuits, mechanical organs are examples that come easily to mind. A good example of bioengineering you can relate to is beer making (yeast engineering in disguise). However, over the last few years, two fields -developmental and stem cell biology- have been coming together unknowingly with a common nexus through Engineering, a new kind of Engineering that was the motivation of the Pasteur meeting. Let me explain.
It is about five years that the tragically late Yoshiki Sasai surprised everybody by showing that Embryonic Stem (ES) cells could be coaxed into forming eye cups and forebrains1,2. His work coincided with the observation that intestinal stem cells would build crypts on their own3 and was followed by a plethora of other reports showing that (but not how) embryonic and adult stem cells can give rise to tissues, organs and structures. These observations are often hailed as the dawn of a new field, organoid biology, though one wonders if this is not, really, Developmental Biology by another name and with one significant difference: embryos, the realm of Developmental Biology, are reproducible, organoids more often than not, aren’t; this has consequences and sets a target.
Sasai was, at heart, a developmental biologist and, having worked with Eddy de Robertis (but probably from before) he was aware of classical experiments in which when animal caps from frogs are left to their own devices, they will make eye cups and forebrains (see Hurtado and de Robertis for a practical review 4). Furthermore, as shown in an insightful review in Development 5 he clearly knew the early development of the eye cup and the lens and thus understood the necessity to relate whatever happens in an ES cell culture to what happens in embryos. Thus he had a good sense for the fact that the autonomous potential of ES cells to organize into tissues autonomously was neither new nor unusual, it reflected what those cells are meant to do. The autonomous gastrulation of frog tissue (exogastrulae), the ability of cells from limb buds to to self organize into digits and the remarkable but much forgotten experiment 6 in which when frog animal cap cells are jumbled up, reagregated and exposed to signals they organize in space as the normal animal caps do, are some examples of this. If you allow me,  ‘organoid biology’ is a rebranding of ‘tissue and organ morphogenesis’ with, perhaps, the added spice that it is making use of our ability to differentiate stem cells rather than the material provided by embryos. It also carries a lot of hype.
The sight of disembodied organ-like structures in culture dishes easily captures the imagination. Probably it is for this reason that the report that a human brain had been grown in a dish 7sparked a huge amount of attention and interest amidst researchers and the public. The press fuelled this interest with headlines such as “Mini brains allow scientists to study brain disorders†or ‘lab grown minibrains aid Alzheimer disease researchers’ that created expectations. What had been achieved did not live up to the billing. What had been observed was not that different from what had been achieved in the mouse system but the fact that it was human, as these things tend to be, and that it had been achieved from iPS cells (genetically engineered ES cells) was an irresistible combination. There was also a sideline about microcephaly and the possibility of modelling brain diseases but one wonders how much of this had to do with journal headline and how much with real science. We certainly look forward to further reports on this front.
There is little question that this was an achievement but what the media forgot to say is that the events that led to those structures are the creations of cells over which we have little control. Furthermore, that in the v1.0 there were no brains as such but rather whimsical structures with a mixture of elements from a brain (forebrain, midbrain and eye tissue), that the system was hardly reproducible, that we had little understanding over what had happened. Feynman famously left on his final blackboard the statement “that which I cannot build, I do not understand’. If in ignorance we extrapolate this to the organoids that grow in culture dishes we would have to say: that which we can build we do not understand. The reason is, of course, that we have not built anything ourselves, that it is the cells -with rules that at the moment we cannot fathom- that have done it (as they are programmed to do), that we are at the mercy of the cells, privileged spectators of their productions. Can we change this? What is missing? Enter the engineers.
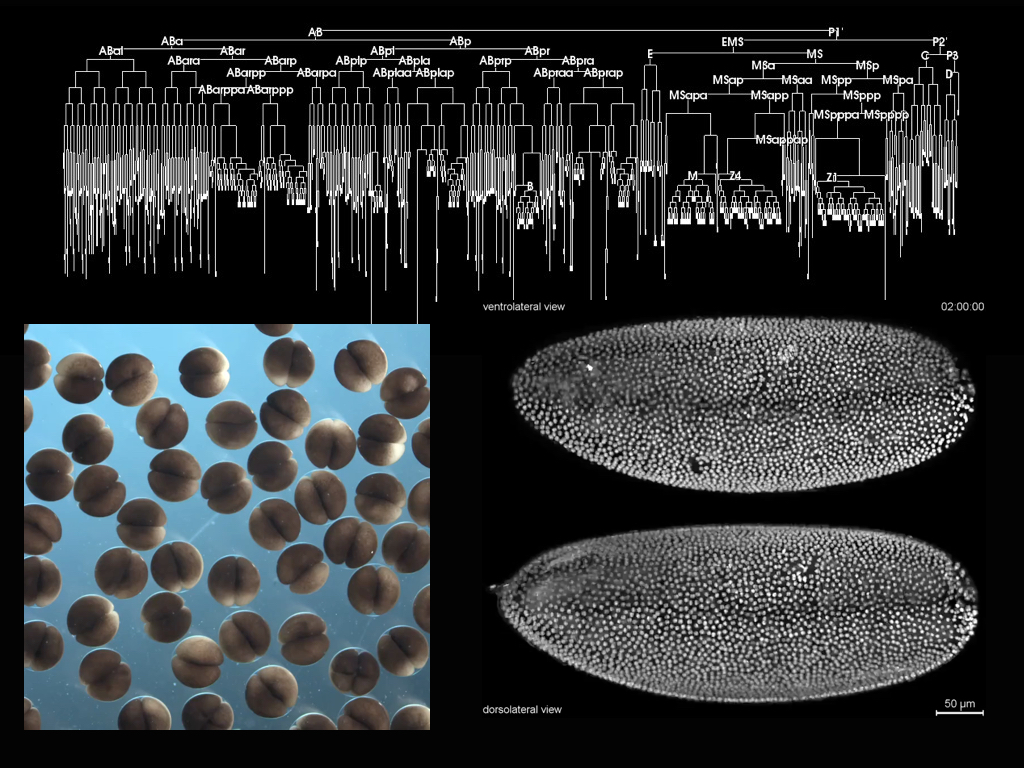 If the system was engineered it would be reproducible and this would allow us to learn. Reproducibility is a first and most important target of the organoid game at this stage and it is where the interaction with engineers is key. Embryos are reproducible systems and it is because they are reproducible that we can use them to learn, that we can detect small changes in patterns to obtain clues of how genes relate to the building of tissues and organs. As F. Jacob pointed out, organisms are the result of evolutionary tinkering, bricolage, a more rudimentary form of engineering and so, perhaps, a way to understand them is to look at them from the point of view of engineering. The goal is not to GUESS how an organism is made, BUT to KNOW how it is made and for this only if we obtain reproducibility shall we understand. If we want to use organoids, we need to strive to make them reproducible and then, not only we shall be able to use them but, they will also teach us about the processes and interests that they represent. Recently there has been some progress in some of the organoid cases and the possibilities are clear. Thus an engineering of the minibrain system has allowed insights into the working of the Zika virus and promises more 8,9, and an engineering of the already robust intestinal organoids, has created the conditions for a sophisticated degree of reproducibility that will contribute to ongoing studies10. These are examples to follow because for the most part the field (if we admit that it is a field) remains hostage to the vagaries of the interactions between cells and culture.
If the system was engineered it would be reproducible and this would allow us to learn. Reproducibility is a first and most important target of the organoid game at this stage and it is where the interaction with engineers is key. Embryos are reproducible systems and it is because they are reproducible that we can use them to learn, that we can detect small changes in patterns to obtain clues of how genes relate to the building of tissues and organs. As F. Jacob pointed out, organisms are the result of evolutionary tinkering, bricolage, a more rudimentary form of engineering and so, perhaps, a way to understand them is to look at them from the point of view of engineering. The goal is not to GUESS how an organism is made, BUT to KNOW how it is made and for this only if we obtain reproducibility shall we understand. If we want to use organoids, we need to strive to make them reproducible and then, not only we shall be able to use them but, they will also teach us about the processes and interests that they represent. Recently there has been some progress in some of the organoid cases and the possibilities are clear. Thus an engineering of the minibrain system has allowed insights into the working of the Zika virus and promises more 8,9, and an engineering of the already robust intestinal organoids, has created the conditions for a sophisticated degree of reproducibility that will contribute to ongoing studies10. These are examples to follow because for the most part the field (if we admit that it is a field) remains hostage to the vagaries of the interactions between cells and culture.
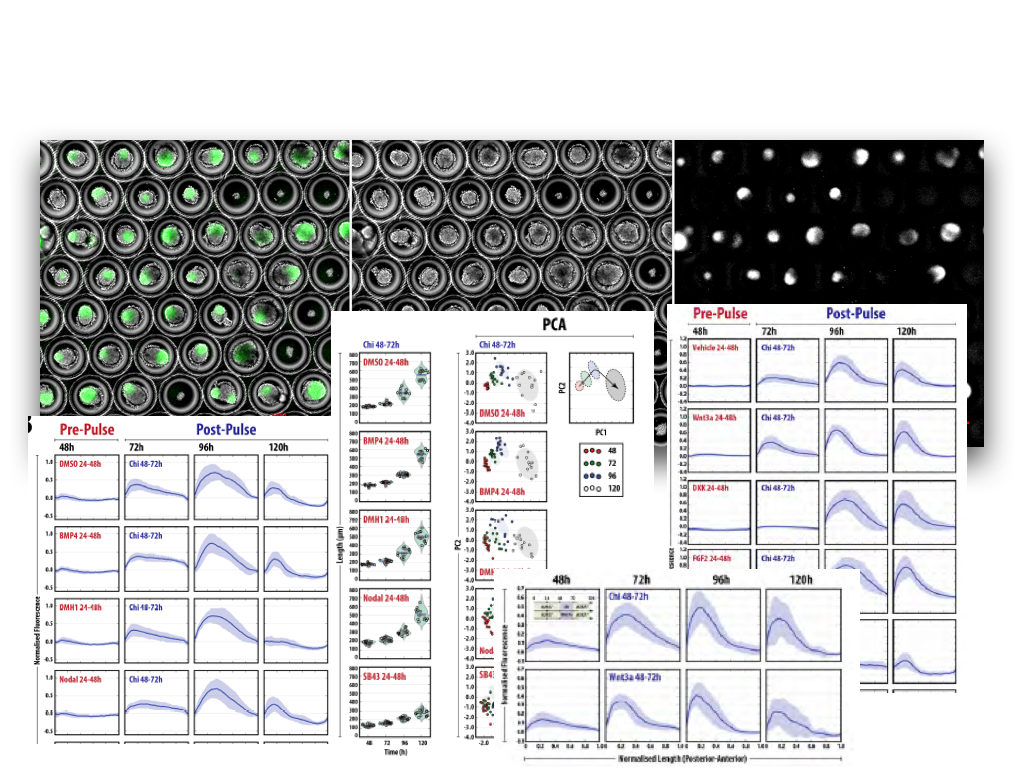 If we agree that the solution is to engineer the process, we need to use the Physics of the system, which, like in any engineering process, underpins the events. This means that in our attempt to engage cells into building tissues and organs we need to engage developmental biologist. If stem cells are the materials, developmental biology is to organoids what Physics is to Engineering, and we need to use it like that. And by Developmental Biology I do not simply mean the garden variety that dabbles in genes and cells but the more quantitative systems one that strives to integrate the rapidly emerging data into models that identifies parameters and tells us how the different variables interact. Just like Evolution tinkers, we shall tinker. At this, as Peter Zandstra exhorted us at the meeting, we need to use what we know in a realistic manner. Build models that contain real dimensions and time scales and use them, like engineers do, to improve, to build, to understand.
If we agree that the solution is to engineer the process, we need to use the Physics of the system, which, like in any engineering process, underpins the events. This means that in our attempt to engage cells into building tissues and organs we need to engage developmental biologist. If stem cells are the materials, developmental biology is to organoids what Physics is to Engineering, and we need to use it like that. And by Developmental Biology I do not simply mean the garden variety that dabbles in genes and cells but the more quantitative systems one that strives to integrate the rapidly emerging data into models that identifies parameters and tells us how the different variables interact. Just like Evolution tinkers, we shall tinker. At this, as Peter Zandstra exhorted us at the meeting, we need to use what we know in a realistic manner. Build models that contain real dimensions and time scales and use them, like engineers do, to improve, to build, to understand.
Organoids, in its many varieties (embryonic and adult stem cells, micropatterned cell arrays, scaffolding of cells from different tissues) promise much but we need to accept that the process is more important than the end, that understanding their beginnings and how they unfold will allow us to understand and improve the end product. One intriguing feature emerging from the current studies and that we shall have to address, concerns the differences between the events that we see in the dish and what we observe in an embryo (see refs 5 and 11 for discussions). It is too early to say whether these differences are important but, it might be that as in engineering there are many ways to make a bridge or a jet or a computer, the same might apply to a tissue and an organ; to put it clearly, there are many molecular solutions to an organ or a tissue and the embryo uses one but we might be able to use others that are simpler. Much interesting and unknown lies ahead but one of the most exciting prospects is the possibility to explore human developmental biology and, in the end, to create a proper organ and tissue engineering as a first step for a regenerative medicine. But we should not cut corners.
Much of this was discussed on and off stage at the Pasteur meeting which had a sense of first encounter between engineers, developmental and stem cell biologists and of expectation of what can be achieved by working together. There will be a report of the meeting in the Development journal, but importantly interesting things will happen. Stay tuned.
1.        Eiraku, M. et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519-32 (2008).
2.        Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51-6 (2011).
3.        Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262-5 (2009).
4.        Hurtado, C. & De Robertis, E.M. Neural induction in the absence of organizer in salamanders is mediated by MAPK. Dev Biol 307, 282-9 (2007).
5.        Sasai, Y., Eiraku, M. & Suga, H. In vitro organogenesis in three dimensions: self-organising stem cells. Development 139, 4111-21 (2012).
6.        Green, J.B., Dominguez, I. & Davidson, L.A. Self-organization of vertebrate mesoderm based on simple boundary conditions. Dev Dyn 231, 576-81 (2004).
7.        Lancaster, M.A. & Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 9, 2329-40 (2014).
8.        Qian, X. et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238-54 (2016).
9.        Xu, M. et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 22, 1101-1107 (2016).
10.      Gjorevski, N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560-564 (2016).
11.      Turner, D.A., Baillie-Johnson, P. & Martinez Arias, A. Organoids and the genetically encoded self-assembly of embryonic stem cells. Bioessays 38, 181-91 (2016).

Very nice post, I’ve enjoyed some of your others as well.
“If stem cells are the materials, developmental biology is to organoids what Physics is to Engineering, and we need to use it like that.”
As a developmental biologist turned engineer, I cannot agree with this statement more. Hopefully more engineers and biologists catch on to this concept.
P.S. – I very much liked your lab’s recent review on genetically encoded self assembly. I think it’s important to make the genetically encoded distinction when discussing organoids.
You are preaching to the choir here; having worked with organoid (or 3D stem cell) culture, and also having read the many M. Lancaster reviews (+ others) it has become very obvious that the only way of (truly) reducing variability within stem cell engineering of tissues is to simply LEARN more about the original tissue/s. This presents a remarkable conundrum, as far as I can see, people simply haven’t investigated the neurodevelopmental niche of humans, for obvious reasons. But we also cannot depend upon animal studies, as it has already been shown the development of humans and our closest relatives differs significantly due to the presence of expanded progenitor populations. In a nutshell, we have currently gone as simple as we can go – we have got to the point where we let the cells do what needs must to develop a tissue. From here on out we must add increasingly complicated instructions (perhaps in a non-complicated delivery) in the aim of developing a ‘true’ model of the human brain. I applaud you on the article, I have never seen the issue put so concisely.